About
Hi, my name is Erin. I'm a scientist based in California's Bay Area and Central Valley.
My expertise lies at the intersection of microbiology, immunology, and genetics alongside a growing interest in plant biology and pathology.
I’m currently a Postdoc and Schmidt Science Fellow at the University of California, Davis
advised by Dr. Pam Ronald.
Here, I'm studying new immune genes or pathways in plants that are conserved across all domains of life.
I aim to use this research to develop biological strategies that boost crop resistant to biotic stresses.
I recently received my PhD from the Biomedical Sciences Graduate Program at the University of California, San Francisco.
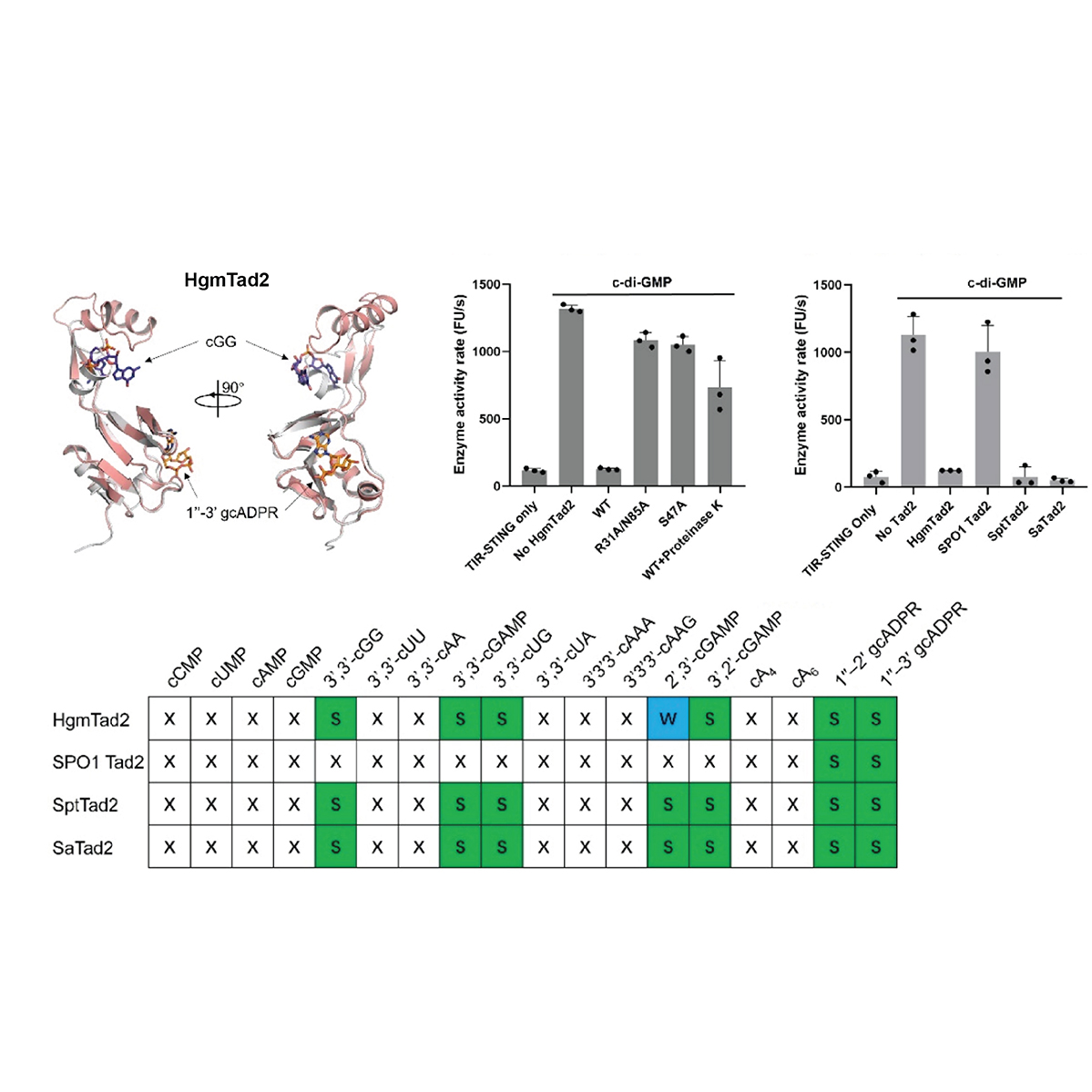
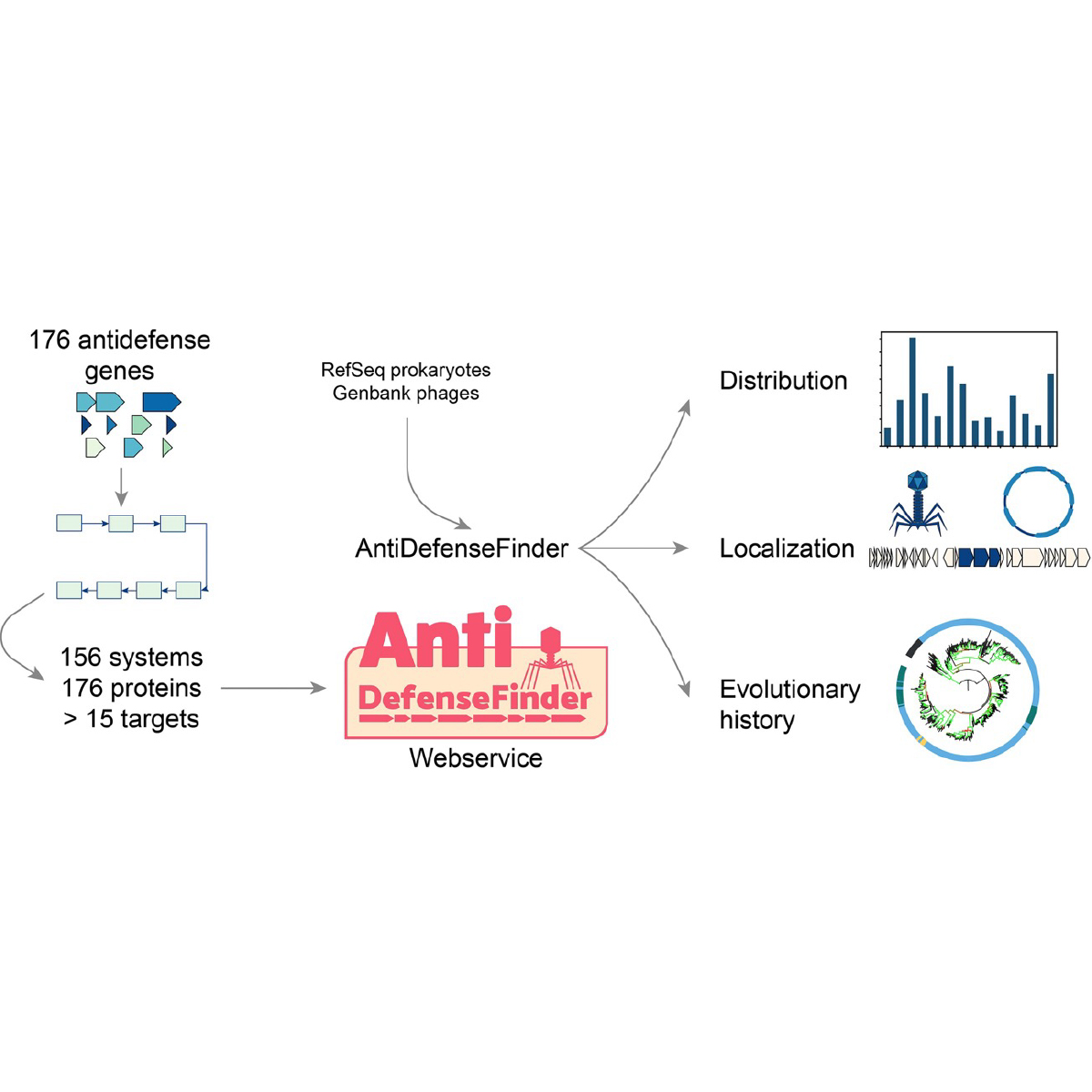
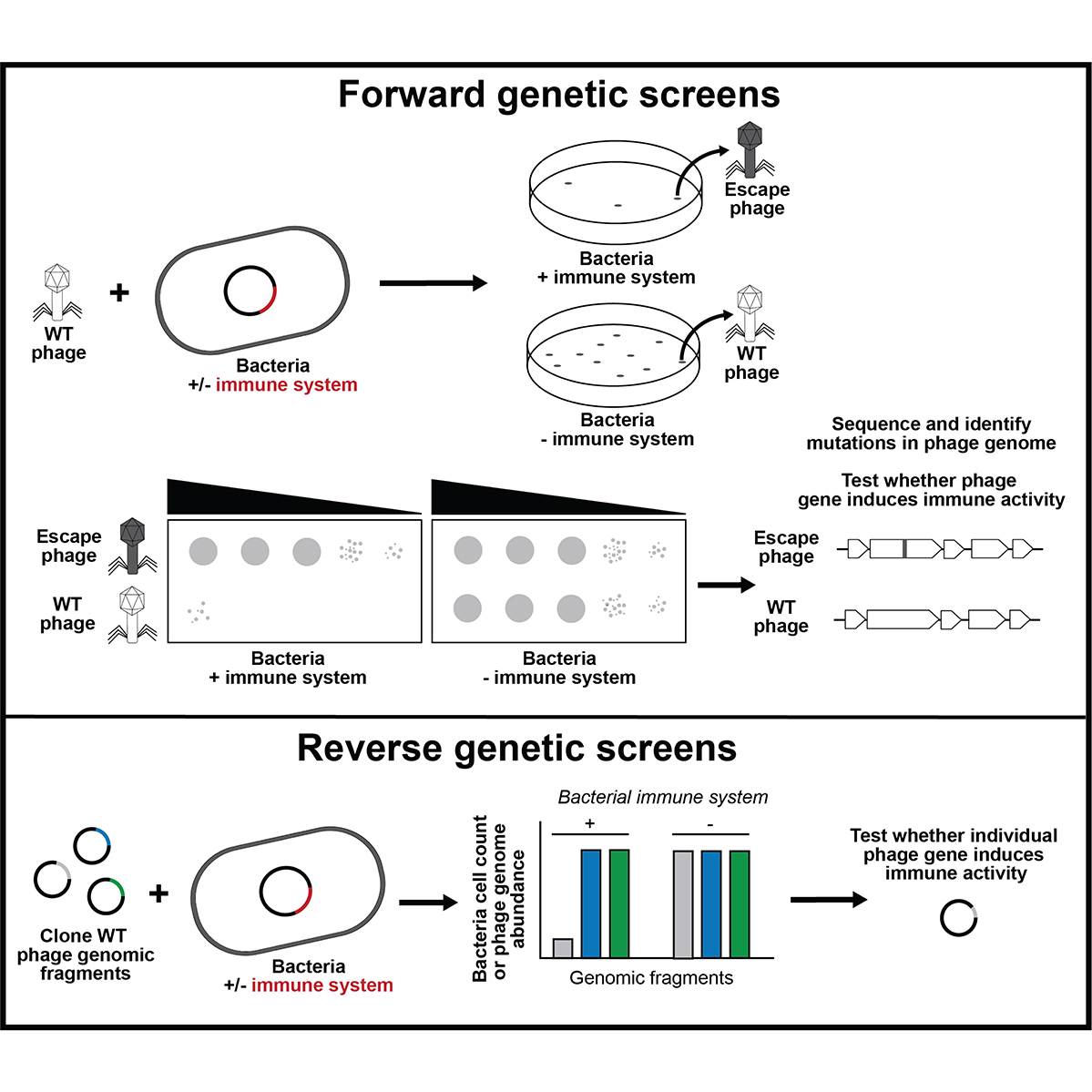
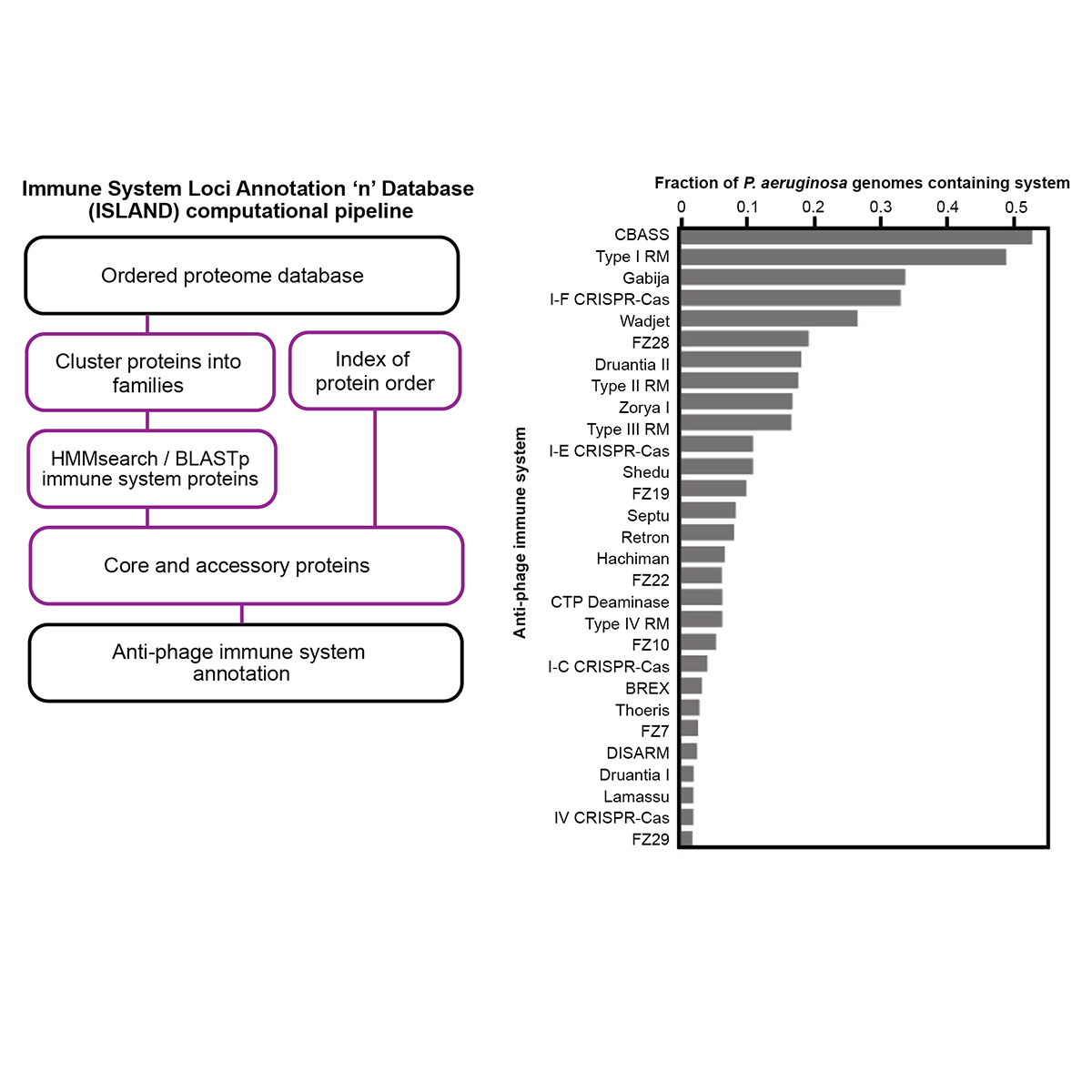
Working in the lab of Dr. Joe Bondy-Denomy, I studied how new bacterial ‘immune systems’ work and co-evolve with bacteriophage (phage)
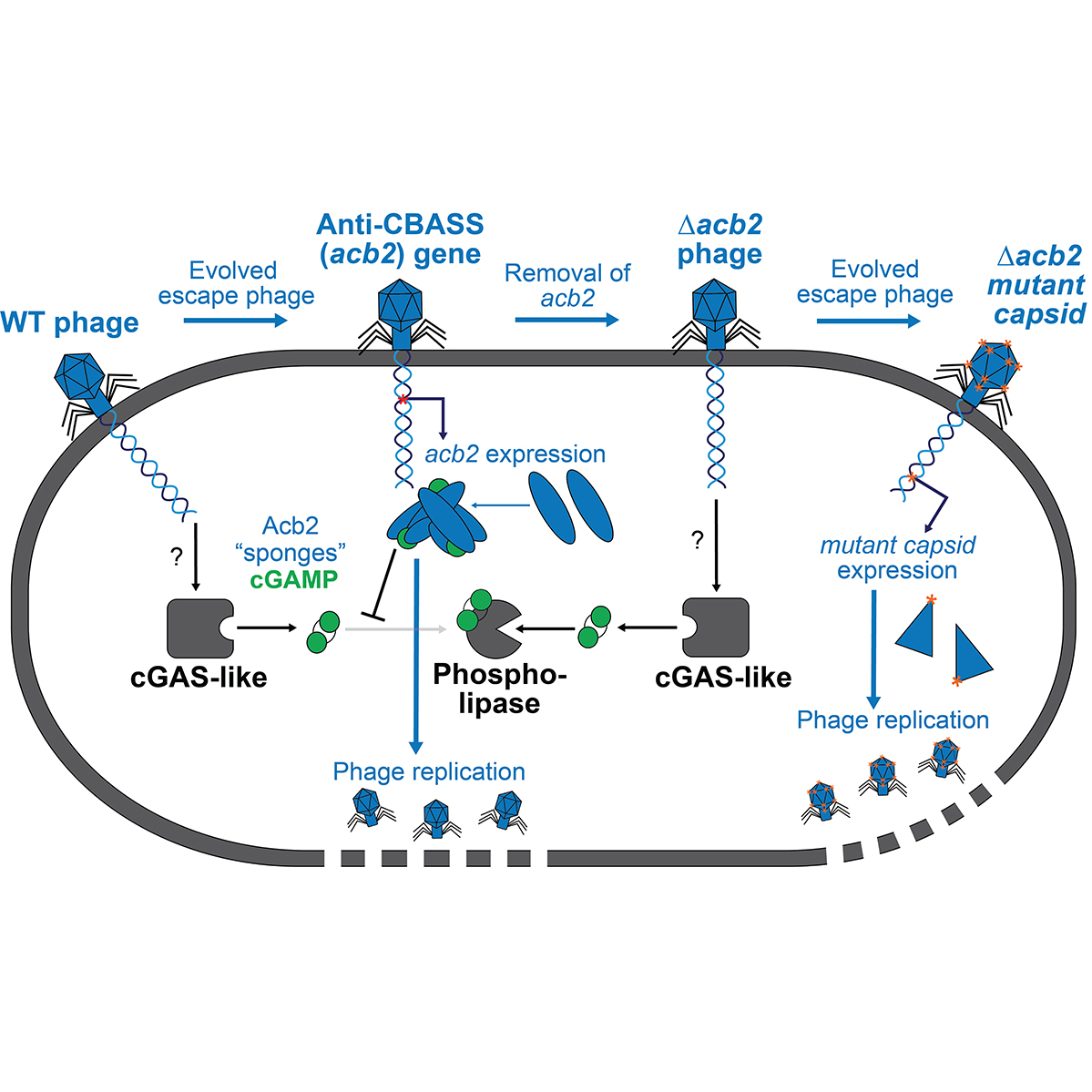
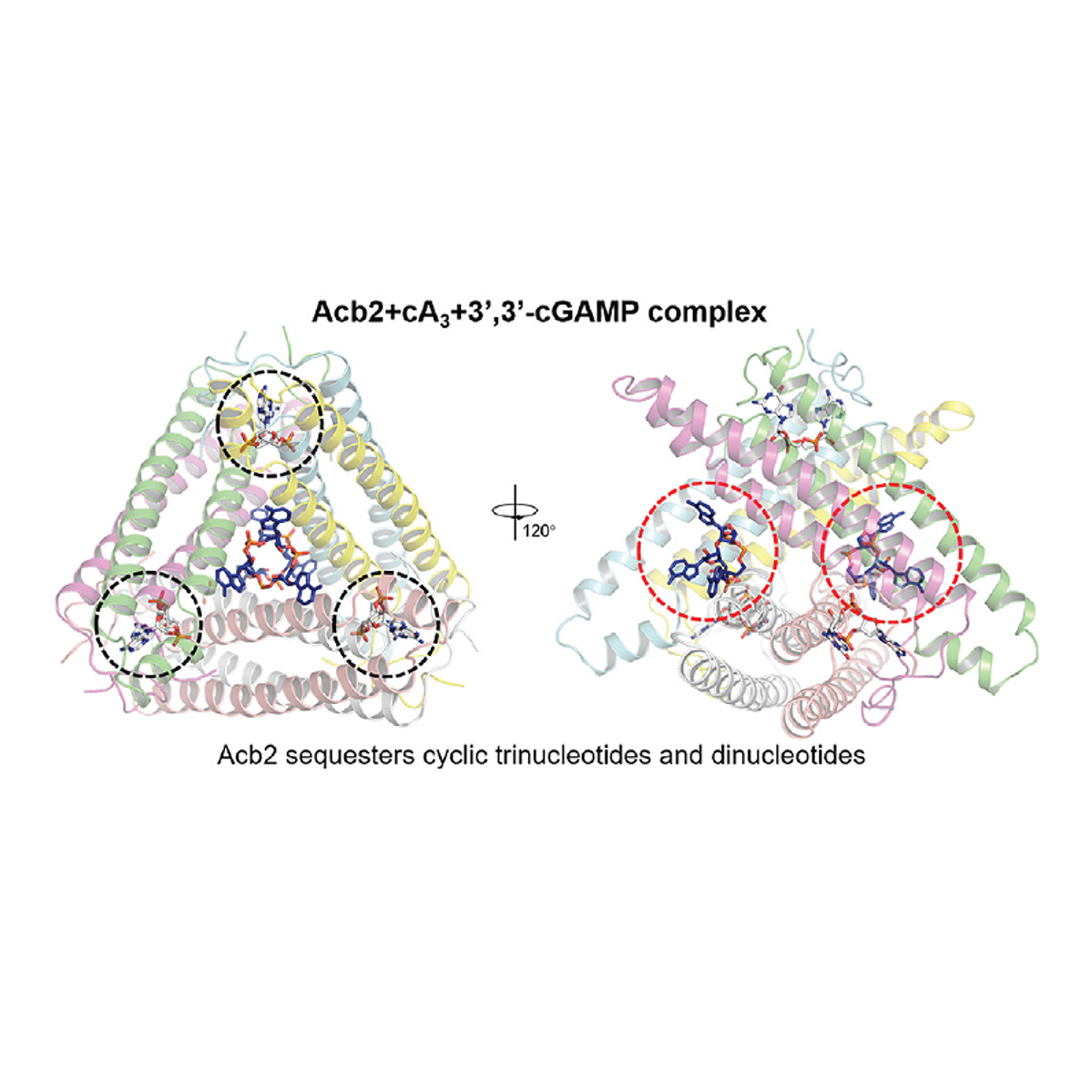
using classic microbiology approaches coupled with modern genome engineering tools. Here, I discovered a viral 'sponge' protein that sequesters many unique immune signals, including cGAMP, that is present in immune systems across all domains of life.
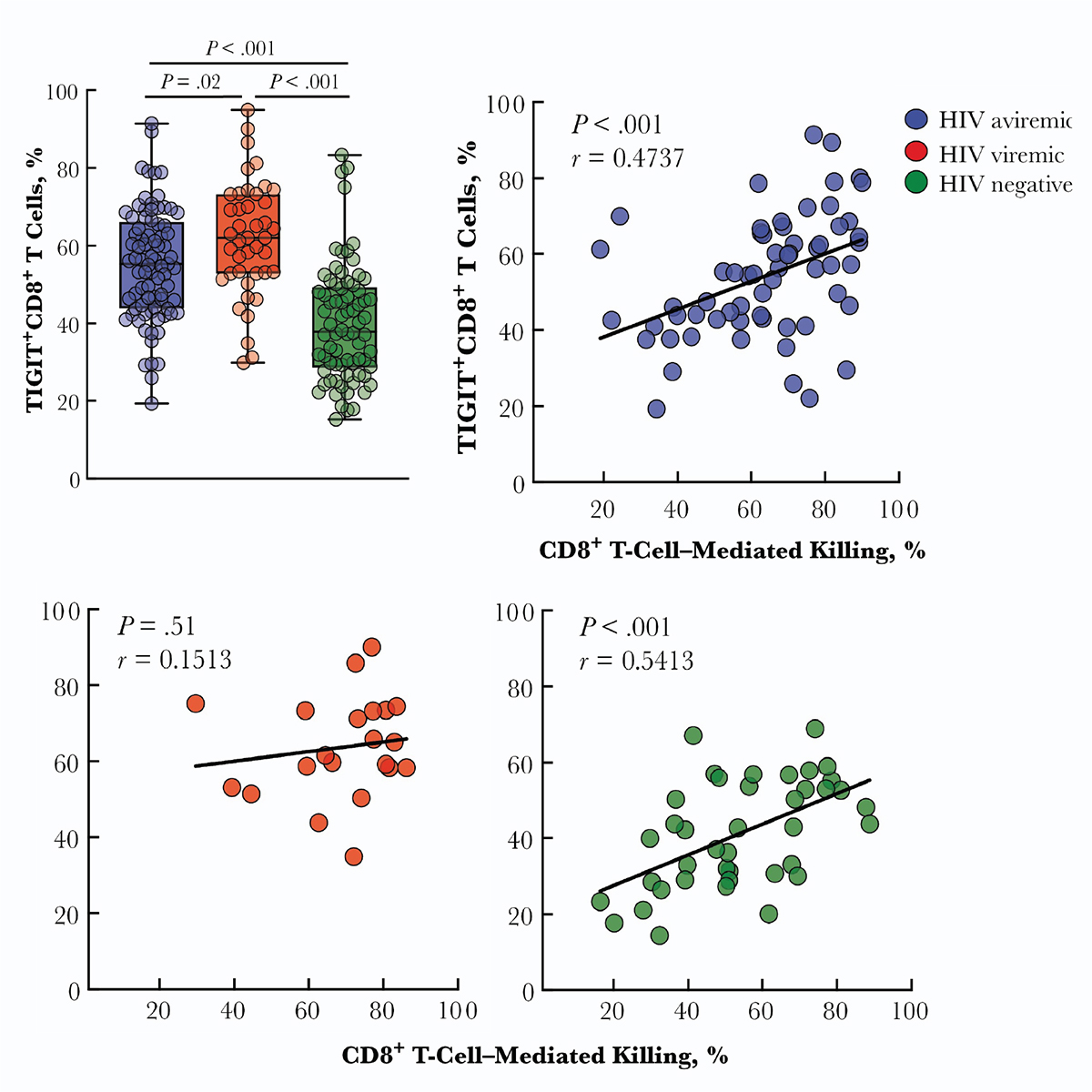
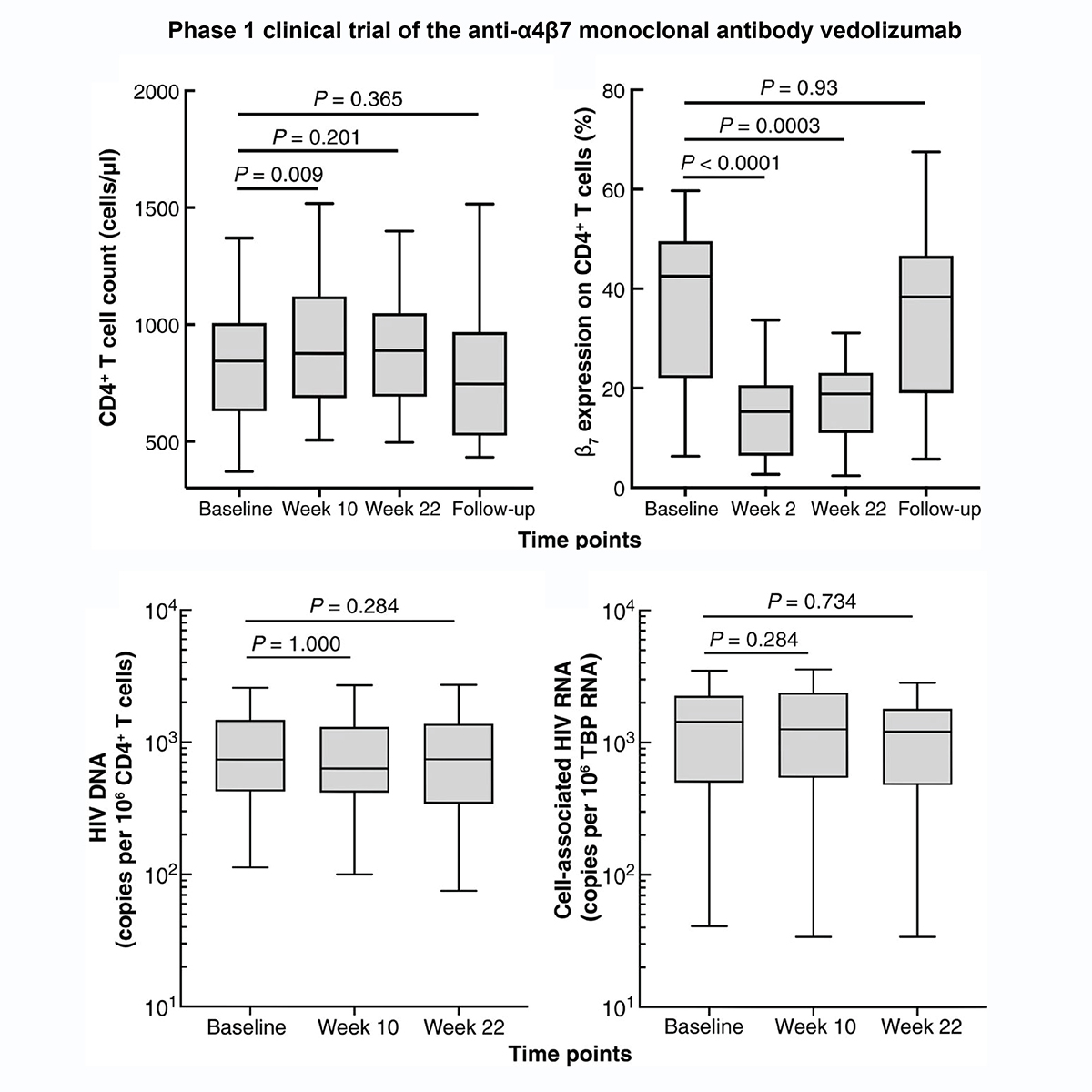
Prior to graduate school, I was at the National Institute of Allergy and Infectious Diseases
as a post-baccalaureate fellow and studied HIV immunopathology and efficacy of HIV therapeutics.
Beyond science, I’m a committed advocate of gender equity and a Director of the
organization Immunologists for Gender Equity
and a Board Member of Women's Global Empowerment Fund.
When I’m not doing science or advocacy work,
I’m an avid reader, coffee consumer, and (moderately above average) rock climber.
If you’d like to get in touch, please send me an email.
I love meeting new people and want to learn more about sustainable food and agricultural practices, as well as
and ways to create innovative and inclusive learning environments.